DNA Fingerprinting also known as DNA Profiling is a technique used by scientists to distinguish between individuals of the same species using only samples of their DNA. Although 99.9% of human DNA sequences are the same in every person, enough of the DNA is different to distinguish one individual from another; unless they are identical (monozygotic) twins, meaning that they develop from one zygote (an egg which has been fertilized by a sperm), which splits and forms two embryos.
|
|
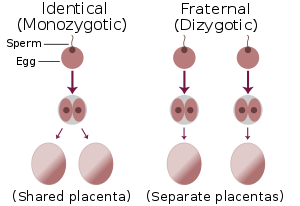
Although this might sound like a small amount, it means that there are around three million base pairs that are different between two people. These differences can be compared and used to help distinguish you from someone else. Only a small sample of cells is needed for DNA fingerprinting. A drop of blood or a root of hair contains enough DNA for testing. Typical biological materials used for DNA profiling includes blood, hair, saliva, semen, and other body tissue cells.
Who invented it?
In 1984, Alec Jeffreys discovered the technique of genetic fingerprinting in a laboratory in the Department of Genetics at the University of Leicester. Colin Pitchfork was the first person in the world to be convicted of murder based on DNA profiling evidence.

Alec Jeffreys
Stages of DNA fingerprinting
Stage 1 Cells are broken down to release DNA
All animal cells are surrounded by a cell membrane and DNA is enclosed within the nucleus. To disrupt the cell membrane and get the DNA into the solution a detergent is used. (Sodium Dodecyl Sulphate). Since DNA in the nucleus of the cell is molded, folded, and protected by proteins; an enzymatic reaction is used break up the protein. For this reaction, Proteinase K is used. Once DNA is purified, alcohol precipitation is done to precipitate DNA. This is done by diluting the DNA with a monovalent salt, adding alcohol to it & mixing gently. 100% Ethanol and Isopropanol are the widely used alcohols for nucleic acid precipitation. Salt neutralizes the charge on the DNA backbone (negatively charged phosphate groups), causing the DNA to become less hydrophilic and fall out of the solution. The precipitated DNA can be pelleted by centrifugation. The salts and alcohol remnants are removed by washing with 70% alcohol. If only a small amount of DNA is available, by Polymerase Chain Reaction, amplified DNA can be obtained. The polymerase chain reaction is a technique used in molecular biology to amplify a single copy or a few copies of a piece of DNA across several orders of magnitude, generating thousands to millions of copies of a particular DNA sequence.
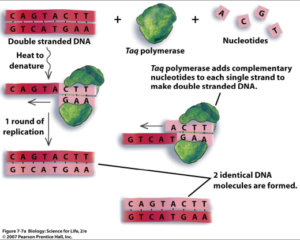
(Figure 2)
Laboratory Manual for Biology: Science for Life, 2nd Edition
Stage 2 DNA is cut into fragments using restriction enzyme
DNA is cut into millions of small fragments with the help of an enzyme called Restriction Endonuclease. Restriction enzymes chop DNA at specific sequences. Like all enzymes, a restriction enzyme works by shape-to-shape matching. When it comes into contact with a DNA sequence with a shape that matches a part of the enzyme, called the recognition site, it wraps around the DNA and causes a break in both strands of the DNA molecule. Each restriction enzyme recognizes a different and specific recognition site, or DNA sequence. Recognition sites are usually only short 4-8 nucleotides. The patterns occur in different places in different individuals and the length of the fragment differs from person to person. Sections of DNA that are cut are called Restriction fragments.
Fragments are separated on the basis of size using a process called “gel electrophoresis”. The DNA is loaded into wells at one end of a porous gel, usually agarose which act a bit like a sieve. An electric current is applied which pull the negatively charged DNA through the gel towards the positive end. The shorter pieces of DNA will move through the gel easily and therefore faster because they don’t need to squeeze through the gels’ pores. It is more difficult for the longer pieces of DNA to move through the gel so they travel slower. As a result, by the time the electric current is switched off, the DNA pieces will be separated in order of size. The smallest DNA molecules will be furthest away from where the original sample was loaded on to the gel. (Figure 4)
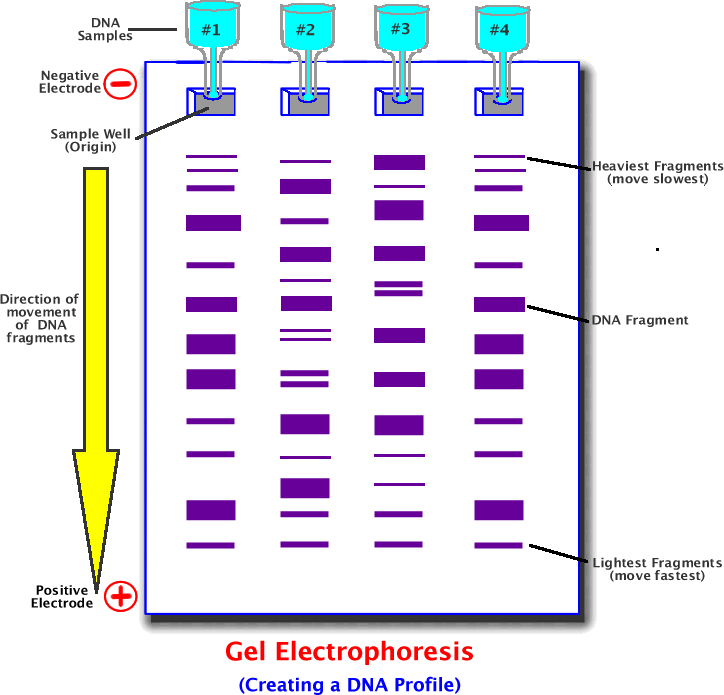
Figure 4
http://science.halleyhosting.com
Stage 4 Southern blotting
Once the DNA is sorted, the pieces of DNA are transferred (referred to as blotting) out of the gel on to a robust piece of nylon or nitrocellulose membrane. Then the membrane is treated with chemicals, usually an alkaline solution, that break the hydrogen bonds between double stranded DNA to make it single stranded. The single stranded DNA is then fixed to the nylon membrane using heat or a UV light and kept overnight.
Stage 5 Probing
After fixing in to the nylon membrane, a probe is introduced. A probe is a radioactive element with a labeled DNA sequence which is used to identify the position of a segment with the complementary sequence by binding to it. DNA probes can also be used to identify the presence of complementary sequences in a mix of fragments. The probe sticks to the fragments of the DNA that has the matching sequence.
The probe shows up on photographic film because the strands of DNA decay and give off light. In the end it leaves dark spots on the film which is also known as the DNA bands of a person. What make up the fingerprint are the unique patterns of bands. Once the filter is exposed to the x-ray film, the radioactive DNA sequences are shown and can be seen with the naked eye. This creates a banding pattern or what we know as DNA fingerprints. This technique is called southern blotting.
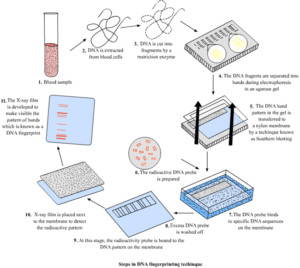
Steps in DNA fingerprinting
Why we use DNA fingerprinting?
- Paternity Testing;
- To find out a person’s parents or siblings and to find out parents of babies who were switched at birth, as Individuals inherit their A variable number tandem repeat (or VNTR) from their parents. A VNTR is a location in a genome where a short nucleotide sequence is organized as a tandem repeat. These can be found on many chromosomes, and often show variations in length between individuals. Each variant acts as an inherited allele, allowing them to be used for personal or parental identification
- Parent child VNTR pattern analysis has been used to solve standard father identification cases
Eg: 1 Who is the father of this child?

- Determining who committed a crime (Forensic science); blood, semen, skin, or other tissue left at the scene of a crime can be analyzed to help prove whether the suspect was or was not present at the crime scene.
- DNA isolated from blood, hair, skin cells, or other genetic evidence left at the crime scene can be compared.
- DNA fingerprints are used to link the suspects using biological evidence
Eg: 2: Who killed Moriarty?


- Identify a body (If the body is badly decomposed/ if only body parts are available after a natural disaster/battle)
- Diagnosis of inherited disorders
- Developing Cures for inherited disorders
Dr. Kaushalya Jayaweera
Reference
https://en.wikipedia.org/wiki/Twin
En.wikipedia.org. (2016). DNA profiling. [online] Available at: https://en.wikipedia.org/wiki/DNA_profiling [Accessed 1 Nov. 2016].
Science.halleyhosting.com. (2016). Gel Electrophoresis. [online] Available at: http://science.halleyhosting.com/sci/ibbio/biotech/electrophoresis.htm [Accessed 1 Nov. 2016].
Www2.le.ac.uk. (2016). A beginner’s guide to DNA fingerprinting — University of Leicester. [online] Available at: http://www2.le.ac.uk/offices/press/for-journalists/code-of-a-killer-1/a-beginners-guide-to-dna-fingerprinting [Accessed 1 Nov. 2016].
